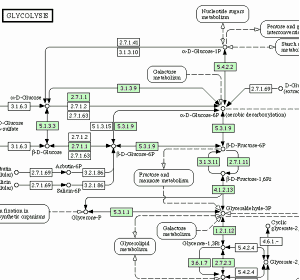
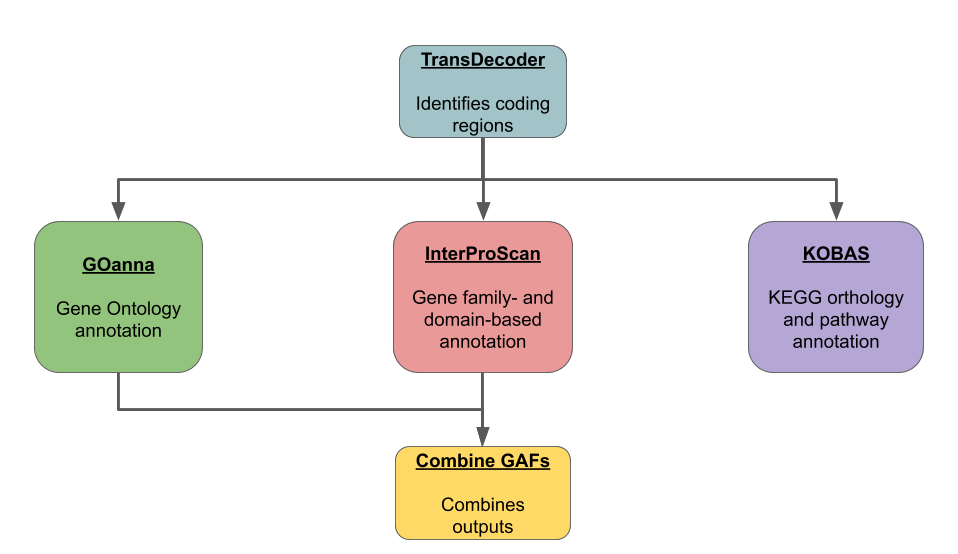
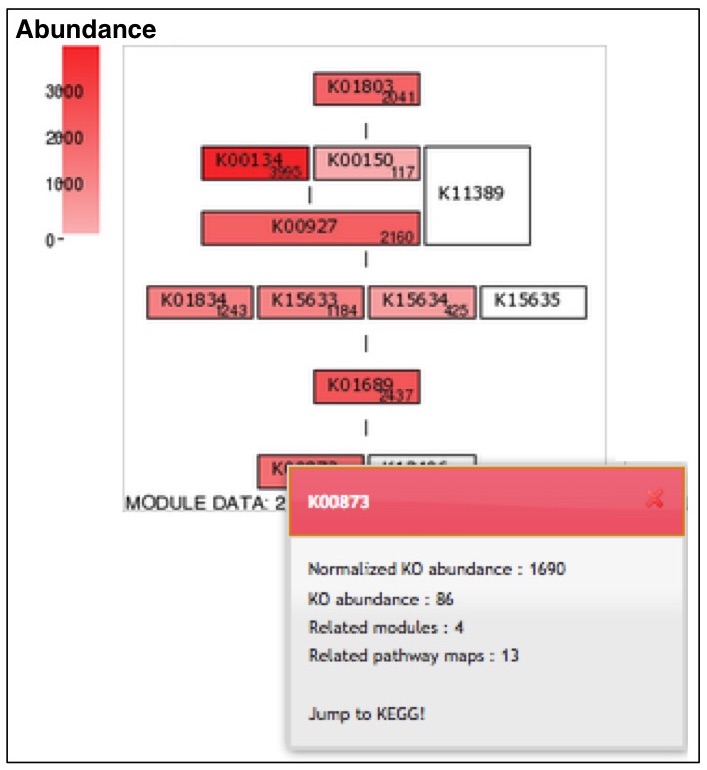
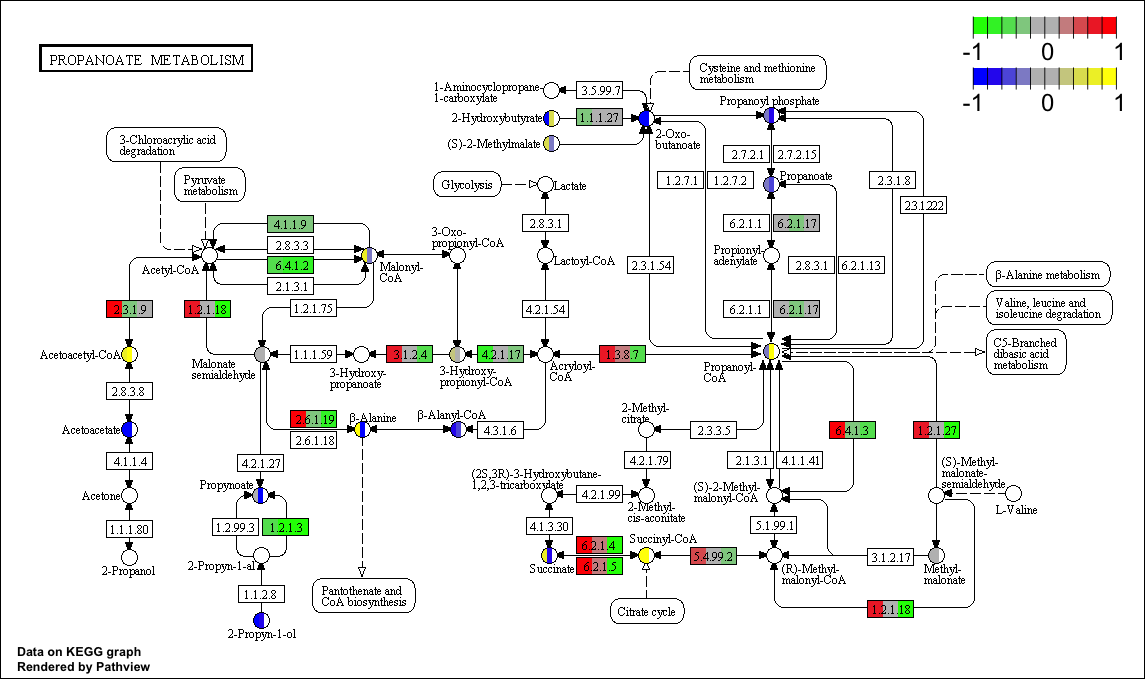
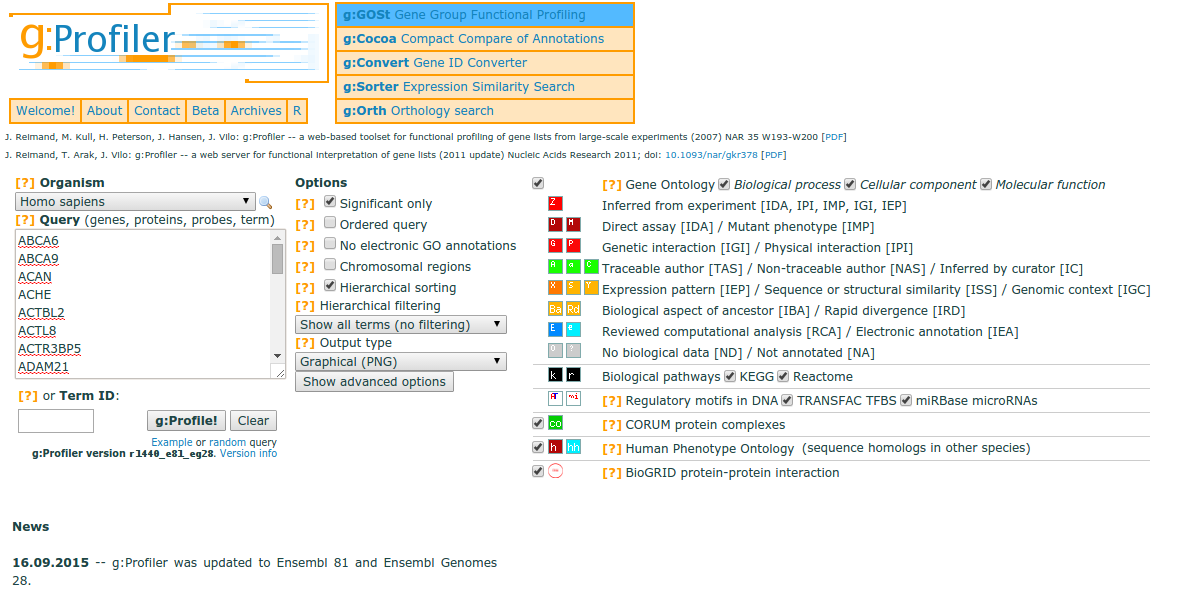
BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences – topic of research paper in Biological sciences. Download scholarly article PDF and read for free on CyberLeninka open
Gene Ontology and KEGG Pathway Enrichment Analysis of a Drug Target-Based Classification System | PLOS ONE

ePath: an online database towards comprehensive essential gene annotation for prokaryotes | Scientific Reports

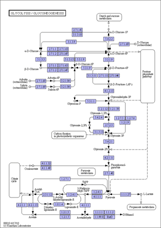
KEGG Mapper for inferring cellular functions from protein sequences - Kanehisa - 2020 - Protein Science - Wiley Online Library

KEMET – A python tool for KEGG Module evaluation and microbial genome annotation expansion - ScienceDirect

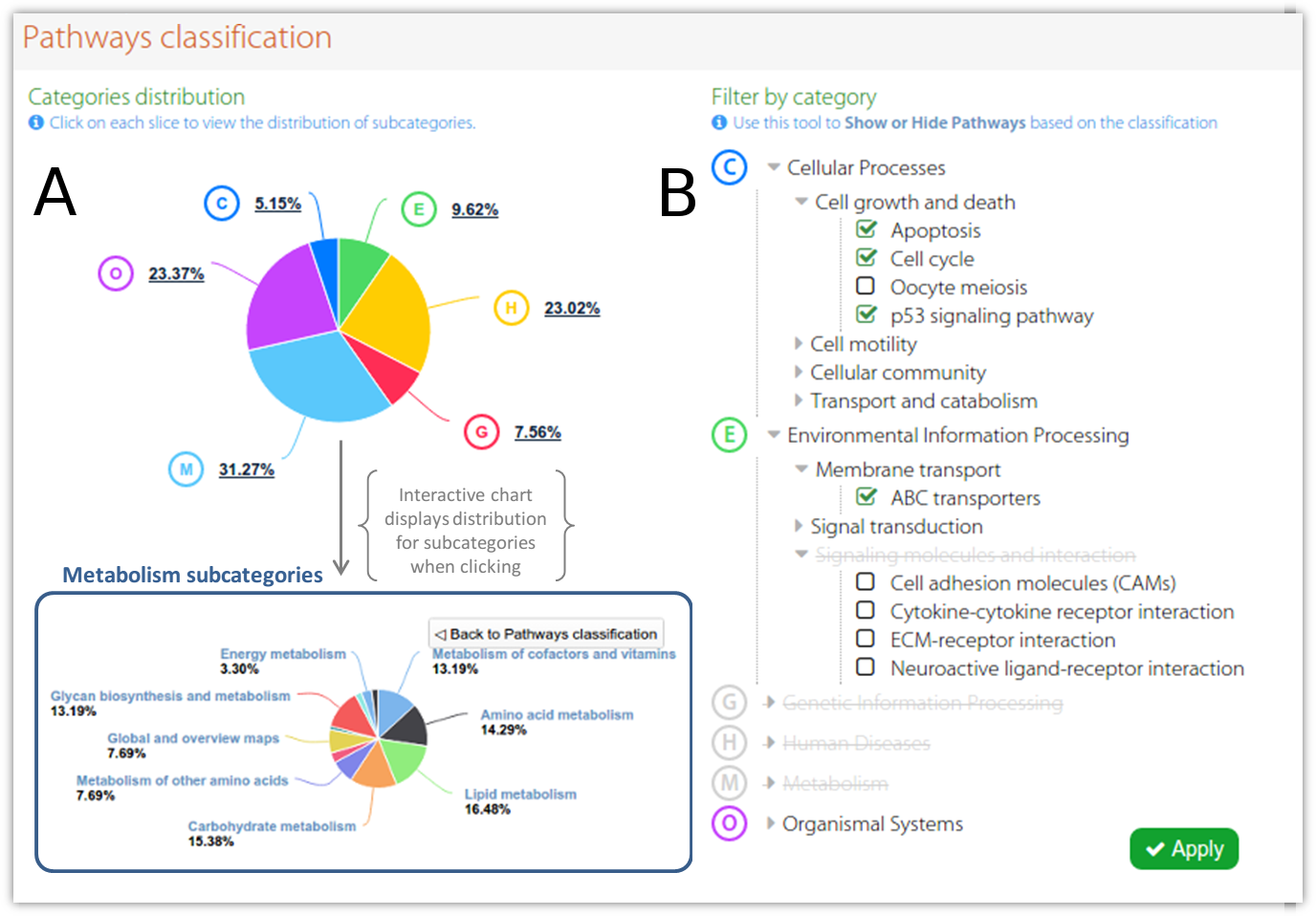
KEGG pathways and GO analysis by DAVID database. (A) GO analysis of... | Download Scientific Diagram

Comparative analysis tools. (i) A pathway is selected from the list of... | Download Scientific Diagram